Author Archives: ap0862
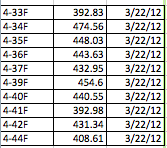
DNAs 4-33 to 4-44 Amplified atpF
Emmanuel and Archana were able to successfully detect regions of DNA from samples numbered 4-33 to 4-44 that were amplified on March 22nd. According to the AASU Genetics Blog, all of the samples (excluding one) yielded a bright band, indicating the presence of a successful amplification of that region. The only sample that was not used was 4-43. After being treated with Exonuclease, the samples that were used were incubated. The Nano-drop spectrophotometer, which measures the quantity of DNA present, was used to measure and quantify our samples so that an accurate result showing the amount of DNA present could be obtained. The whole procedure can be found at https://armstronggenetics3.wordpress.com/category/techniques/. The results, as shown in the data table below, there was a consistently high yield of DNA in all of the samples that were quantified. The first column is the sample number, the second is the quantified number (ng/ul), and the third is the data quantification was performed. The range was 392.83 ng/ul to 474.56 ng/ul, which doesn’t show that much variation in data. As always, there are possible sources of error, such as pipetting, denaturation of DNA, initial mix-up of samples, etc.
Holcoglossum subulifolium
Genus species: Holcoglossum subulifolium, Tribe: Vandeae, Subtribe: Aeridinae, It is a monopodial epiphyte. This means that it produces leaves and flowers from a single stem (monopodial) and grows on another plant for support (epiphyte). Found in the Vietnamese and Chinese montane forests at heights upto 1,600 meters, H. subulifolium produces awl-shaped leaves and white flowers that can grow upto 1 and ½ inches in width. It blooms mainly in the spring and summer.
Optimal growth occurs when they’re planted on tree ferns under full sunlight and high humidity. They have to be consistently watered for successful growth. Sphagnum moss or tree fibers are the best media to grow these orchids on, as they are well- drained and the flowers would thrive.
Sources:
http://www.orchidspecies.com/holsubulifolium.htm
http://orchids.wikia.com/wiki/Holcoglossum_subulifolium
http://www.species-specific.com/view-orchid-species-information.php?id=44



You must be logged in to post a comment.