Category Archives: atpF
DNAs 4-33 to 4-44 Amplified atpF
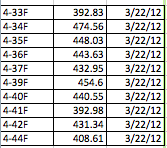
Emmanuel and Archana were able to successfully detect regions of DNA from samples numbered 4-33 to 4-44 that were amplified on March 22nd. According to the AASU Genetics Blog, all of the samples (excluding one) yielded a bright band, indicating the presence of a successful amplification of that region. The only sample that was not used was 4-43. After being treated with Exonuclease, the samples that were used were incubated. The Nano-drop spectrophotometer, which measures the quantity of DNA present, was used to measure and quantify our samples so that an accurate result showing the amount of DNA present could be obtained. The whole procedure can be found at https://armstronggenetics3.wordpress.com/category/techniques/. The results, as shown in the data table below, there was a consistently high yield of DNA in all of the samples that were quantified. The first column is the sample number, the second is the quantified number (ng/ul), and the third is the data quantification was performed. The range was 392.83 ng/ul to 474.56 ng/ul, which doesn’t show that much variation in data. As always, there are possible sources of error, such as pipetting, denaturation of DNA, initial mix-up of samples, etc.
Spilicing of group II introns in spinach chloroplasts (in vivo): analysis of lariat formation
The focus of this research was to analyze the mechanics of chloroplast mRNA splicing in vivo. In order to accomplish this study four RNAs from the spinach chloroplast group II intron-containing genes were analyzed. The genes that were analyzed is as follows atpF, rpoC1, petD, and petB. The researchers did a northern analysis of chloroplast RNA and dectected a lariat-intron 3′ exon splicing intermediates. These RNAs were then treated with a Hela cell debranching extract that made the splicing intermediates disappear and this allowed them to confirm their identities. The lariat splicing intermediates were then further studied by reverse transcriptase to ascertain the branch point location. They found that the in vivo loaction of atpF and petB introns were eight bases upstream of thier known 3′ intron/exon boundaries. In comparison, they did not detect any splicing intermediates by primer extension analysis of petB and rpoC1. the result demonstrated that the amount of lariat- intron 3′ exon-splicing intermediates present in the chloroplast RNA population is less in dealing with rpoC1 and pet B compared to atpF and petD. This to them suggested that the steady-state level of any splicing intermediates is the result of a balance between the splicing kinetics of the particular RNA and sensitivity of its splicing intermediate to degradation. In the end, the researchers concluded that the balance between the two factors varies significantly for chloroplast introns even if the RNA like petB and petD are transcribed from the same promoter.
Reference:
Jeong-Kook, K., and Hollingsworth, M.J. “Spilicing of group II introns in spinach chloroplasts (in vivo): analysis of lariat formation.” Current Genetics. Volume 23 Number 2 (1993),175-180.
Creating a plant DNA barcode using atpF
The chloroplast gene atpF encodes for the ATP synthase subunit CFO I. The gene sequence in this region is highly conserved between members of a species, yet show variability between species. Due to the nature of this sequence, it has been proposed to use the atpF region as a barcode to identification of plant species. Similar techniques are utilized in animals, using mitochondrial sequences for cox1. Plant mitochondria, however, do not include enough variability between species due to the slow evolution of the mitochondrial genome in plants. In the study by Lahaye et. al. 101 taxa were utilized, including 3 Orchid species. The results of this study were indeterminate, and it was suggested that other gene locus be included in the search for a genetic barcode for plants.
Reference: Renaud Lahaye, Vincent Savolainen, Sylvie Duthoit, Olivier Maurin and Michelle van der Bank. 2008. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the Flora of the Kruger National Park as a model system (South Africa). Nature Preceedings.
A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park as a model system
In this study the introns between psbK-pabl and atpF-atpH were tested for use as genetic barcodes in South African monocots. There were a total of 101 species sampled for testing, and the methods where similar to those used in our research although different primers were used. After sequencing and analysis the conclusion was that these introns though helpful when used together are not as accurate as the matK gene.
Reference: Lahaye, R., Savolainen, V., Duthoit, S., Muarin, O., vand der Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park as a model system. (2008) Nature
The subunit b of the F0F1-type ATPase of the bacterium Mycoplasma pneumoniae is a lipoprotein.
In this study, DNA analysis of the F0F1-ATPase operon of the bacterium Mycoplasma pneumoniae indicated that the subunit b, amino acid residue 123 to 207, which is encoded by the gene atpF is a lipoprotein of the murein lipoprotein type of Escherichia coli. The DNA analysis of the F0F1-type ATPase operons of Mycoplasma gallisepticum, Mycoplasma genitalium , and Mycoplasma pneumoniae were compared. They were found to have the greatest stuctural similarities in orthologs and the greatest differences in the component encoded by atpF, the subunit b. Mycoplasma gallisepticum and Mycoplasma genitalium code for conserved lipoprotien consensus sequence whereas all other mycoplasms have characteristic lipoprotein nature of subunit b. From the data observed, an improved model that includes the lipoprotein characteristics of the protein for the procession and arrangement of subunit b was proposed but unfortunately the data did not explain where the lipoprotein pattern of subunit b of the mycoplasmas came from.
Reference:Pyrowolakis, G., Hofmann, D., and Hermann,R. The subunit b of the F0F1-type ATPase of the bacterium Mycoplasma pneumoniae is a lipoprotein. (1998). The Journal of Biological Chemistry.273(38):24792-6
Identifying Escherichia coli genes involved in intrinsic multidrug resistance.
In this study, the source of bacterial resistance to antibiotics was observed through mutations of different genes of Escherichia coli. The wild type E. coli, EC100, was used for comparison. Thousands of genes were mutated to determine which caused more resistance to the antibiotic chloramphenicol. Of the mutations, only thirteen showed significant resistance or susceptibility to the antibiotic. Compared to the wild-type EC100, mutations of the genes rob, garP, bipA, insK, and yhhX caused more susceptibility to chloramphenicol. In comparison, the mutations of the genes rhaB, yejM, dsdX, nagA, yccE, atpF, and htrB caused a higher resistance to chloramphenicol. As a result, mutations of genes such as atpF should be further studied to understand bacterial resistance to antibiotics.
Reference:
Duo, M., Hou, S.,& Ren, D. (2008). Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Applied Microbial Biotechnology, 81, 731-741.
CRS1 is a Novel Group II Intron Splicing Factor that was Derived from a Domain of Ancient Origin
Group II introns are categorized as self-catalytic ribozymes that can perform self-splicing. This splicing can be used to further study the roles that proteins have in facilitating RNA-catalyzed reactions. Genetic screening of the maize genome has revealed two nuclear genes needed to facilitate group II intron splicing in the chloroplast; crs1 and crs2. The research focused on crs1 because researchers believed that this gene facilitates atpF intron splicing. Several reasons have led them to this belief. First, CRS1 has been observed in ribonucleorprotein complexes in the chloroplast with the atpF intron. Second, CRS1 is basic and has a repeated domain with features that suggest a RNA-binding domain. And lastly, the mutated crs1 gene resulted in disrupted splicing of the atpF intron. These findings are used to support the idea that CRS1 is used to influence splicing of the atpF intron by directly binding to it. Also, analysis of the crs1 mutant provides evidence that CRS1 is involved in more than just atpF splicing but also in chloroplast translation.
Reference:
Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. 2001. “CRS1 is a Novel Group II Intron Splicing Factor that was Derived from a Domain of Ancient Origin”. RNA: A Publication of the RNA Society. September 2001. 7: 1227-1238
A Member of the Whirly Family is a multifunctional RNA- and DNA- Binding Protein that is Essential for Chloroplast Biogenisis
This study examines the protein ZmWHY1 and its importance in the promotion of atpF intron splicing in vitro. It also disproves the argument that ZmWHY1 only interacts with DNA and shows its interaction with RNA as well. CRS1 is a known agent in the splicing of an atpF intron; however, it is not efficient enough to do the job on its own, other proteins must be involved. Mass spectrometry (a method of identifying chemicals in a substance based on their mass and charge) was used to identify the other proteins involved in the process of splicing an atpF intron. At the conclusion of this process, ZmWHY1 was found to be one of CRS1 protein’s aids in the splicing process. Observations of disruption between ZmWHY1 and CRS1 in the presence of DNAse or RNAse led the researchers to the conclusion that ZmWHY1 interacts with both DNA and RNA. Further investigation showed that the atpF intron RNA was most attracted by the RNA of ZmWHY1.
References:
Prikryl, J., Watkins, K., Frisco, G., Wijk, K., Barkan, A. 2008. Nucleic Acids Research 36:16.

You must be logged in to post a comment.